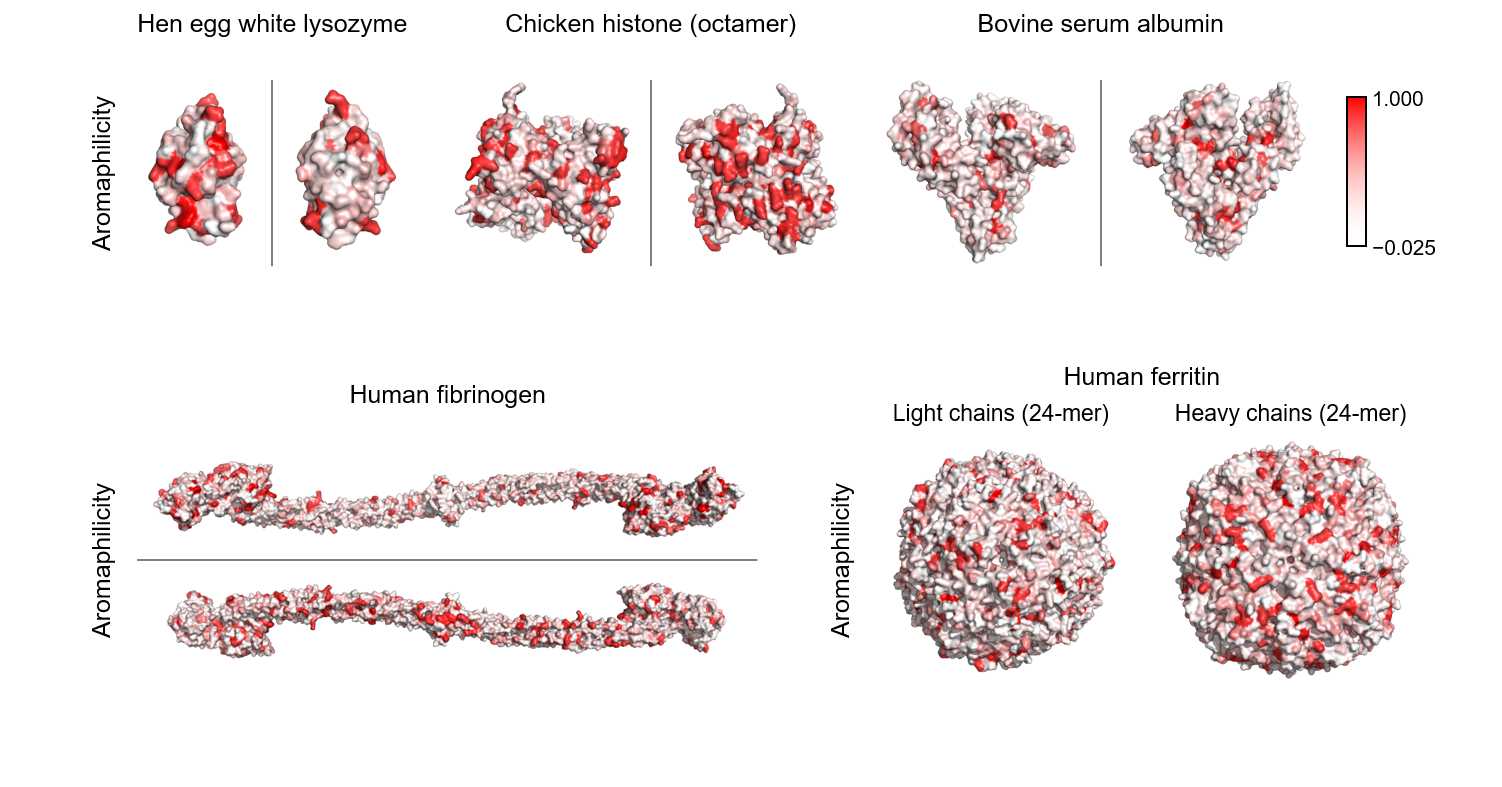Aromaphilicity index
If you use the aromaphilicity index or the pymol script for aromaphilicity mapping, please include an appropriate citation: Atsushi Hirano and Tomoshi Kameda. Aromaphilicity Index of Amino Acids: Molecular Dynamics Simulations of the Protein Binding Affinity for Carbon Nanomaterials. ACS Applied Nano Materials (DOI:10.1021/acsanm.0c03047).
| Amino acid | Aromaphilicity index |
|---|---|
| Trp | 1.000 |
| Tyr | 0.850 |
| Arg | 0.750 |
| Phe | 0.575 |
| His | 0.450 |
| Met | 0.325 |
| Gln | 0.300 |
| Asn | 0.200 |
| Ile | 0.200 |
| Cys | 0.200 |
| Val | 0.150 |
| Ser | 0.125 |
| Leu | 0.125 |
| Pro | 0.125 |
| Lys | 0.100 |
| Thr | 0.075 |
| Glu | 0.050 |
| Ala | 0.025 |
| Gly | 0.000 |
| Asp | -0.025 |
Pymol Script for Aromaphilicity Mapping.
1. Save the script as "aromaphilicity.py".
2. 2.1. Open Pymol file of interest.
2.2. Select 'File'-'Run Script'-aromaphilicity.py
2.3. Write "aromaphilicity" on the command line.
2.4. Show-surface