Registration(on image) Demo Registration (in 3D Space) Demo
Registration of 3D cerebral vessels to X-ray angiograms
Collaborators:J. A. Noble、D.L.Wilson(Oxford Univ.)
Abstract
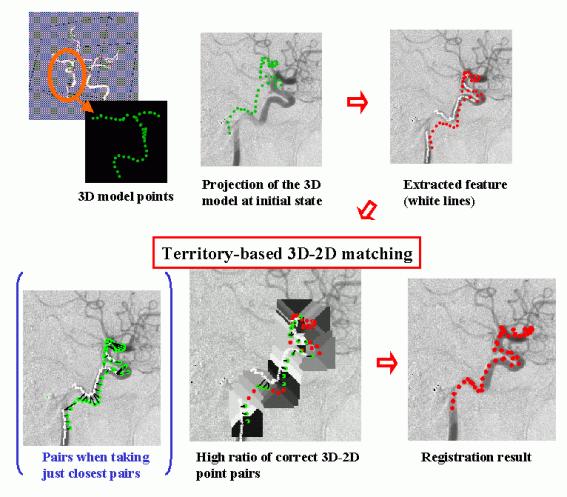
We have developed a quick method to obtain the 3D transformation of a 3D free-form shape model from its 2D projection data in collaboration with the Medical Vision Lab. of the University of Oxford. This method has been developed for the real-time registration of a 3D model of a cerebral vessel tree, obtained from pre-operative data (eg. MR Angiogram), to a X-ray image of the vessel (eg.Digital Subtraction Angiogram) taken during an operation. First, the skeleton of the vessel in a 2D image is automatically extracted in a model-based way using a 2D projection of a 3D model skeleton at the initial state (up to +-20 degree difference in rotation). Corresponding pairs of points on the 3D skeleton and points on the 2D skeleton are determined based on the 2D Euclidean distance between the projection of the model skeleton and the observed skeleton. In the process, an adaptive search region for each model point, which is determined according to the projected shape, effectively removes incorrect correspondences. Based on a good ratio of correct pairs, linearization of a rotation matrix can be used to rapidly calculate the 3D transformation of the model which produces the 2D observed projection.
Related papers
1) Y. Kita, D. L. Wilson, J. A. Noble and N. Kita: "A quick 3D-2D registration method for a wide-range of applications", Proc. of 15th International Conference on Pattern Recognition, pp.981-986, 2000.
2)Y. Kita, D. L. Wilson and J. A. Noble: "Real-time registration of 3D cerebral vessels to X-ray angiograms", Proc. of MICCAI98(1st International Conference on Medical Image Computing and Computer-Assisted Intervention), pp. 1125-1133, 1998.